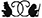A fundamental property that sets life apart from non-life is the ability to sense and react to changes in the environment. While we know that each and every cell must somehow process information about the world around it, it is often difficult to understand why a given collection of biological parts is well suited for a particular task. How do bacteria know where to find food? How does a stem cell know whether to become a bone or muscle cell? A guiding goal of systems biology is to understand the architecture and design of cellular processes that have been shaped by evolution over millions of years.
We sought to study a particular instance of elegant design in biology, that of logarithmic sensing. This phenomenon, where a cell’s response is proportional to the ratio of change in signal as opposed to the absolute difference, is observed across a wide spectrum of biological contexts. This is most apparent in human sensory systems, where we measure our perceived intensity of sound in the logarithmic unit of decibels and our visual system is more sensitive to contrast than to absolute changes in brightness. Essentially we wanted to understand how this sort of behavior, observed in both large-scale cognitive system and in the response of single cells, is encoded at the molecular level.
We found that a large class of proteins, those that are allosterically regulated, are particularly well suited to perform logarithmic sensing. Allosteric proteins are biological molecules that have two or more different conformational states, each of which have a different biological function. A classical example is hemoglobin, the oxygen transport molecule found in almost all vertebrate life. In 1904, Christian Bohr (father of Niels Bohr) observed that hemoglobin would change its affinity for oxygen when he varied the amount of carbon dioxide in the medium. It turns out that high levels of CO2 would bias the hemoglobin towards a low affinity state, and low levels would bias it towards a high affinity state.
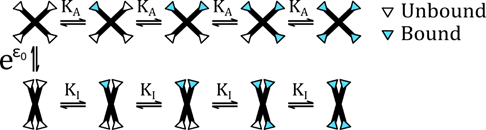
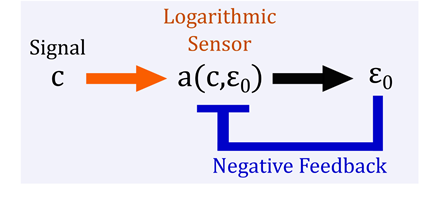
It turns out that the way in which the binding kinetics of allosteric proteins shifts in response to effectors, like CO2 for hemoglobin, is naturally logarithmic. That is to say the activation curve has a regime that looks linear when plotted on a logarithmic scale and can be shifted without changing its overall shape. We show that this behavior fits nicely into a known architecture for Fold-Change Detection, where a logarithmic transformation is coupled with negative feedback.
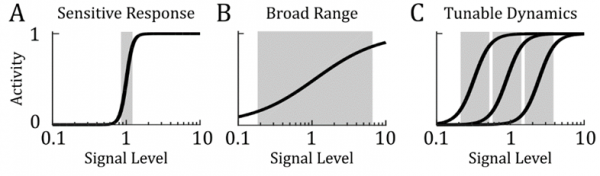
More generally, this work shows how we can think about the biophysical properties of proteins as functional components of sophisticated information processing systems. This moves towards a goal of unifying the low-level properties of molecules with the high-level properties of living systems.
— Noah Olsman, 2017
Reference: Allosteric proteins as logarithmic sensors, PNAS 113 (30): E4423-4430, 2016